What is Eugene?
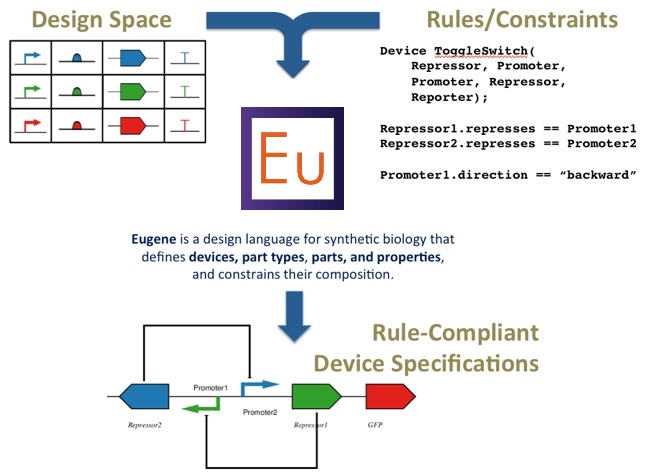
Eugene is an ecosystem of human-readable and machine-executable languages developed to speed up and innovate the design process of synthetic biology at various levels of abstraction, such as Sequences, Parts, Devices, and Systems. Eugene contributes towards standardized languages for communicating Synthetic Biology.
Eugene's facilities for synthetic biology include:
- Specification of (synthetic) biological components
- Support of Data Exchange standards
- Library Management of design and components
- Specification of design constraints ("Rules") on composing components into novel systems
- Built-In functions to enumerate rule-compliant designs automatically
- Control structures and functions for the specification of design synthesis
Publications
E. Oberortner, D. Densmore
miniEugene: Constraint-based Design of Synthetic Biological Systems
ACS Synthetic Biology - Technical Note
DOI:10.1021/sb500352b
E. Oberortner, S. Bhatia, E. Lindgren, D. Densmore
A Rule-based Design Specification Language for Synthetic Biology
ACM Journal on Emerging Technologies in Computing Systems (JETC)
Special Issue on Synthetic Biology
DOI:10.1145/2641571
H. Huang, E. Oberortner, D. Densmore, and A. Kuchinsky
Eugene’s Enriched Set of Features to Design Synthetic Biological Devices
International Workshop on Bio-Design Automation (IWBDA), 2012.
D. Densmore, J. T. Kittleson, L. Bilitchenko, A. Liu, and J. C. Anderson
Rule based constraints for the construction of genetic devices
Proceedings of 2010 IEEE International Symposium on Circuits and Systems (ISCAS), 2010.
DOI:10.1109/ISCAS.2010.5537540
L. Bilitchenko, A. Liu, S. Cheung, E. Weeding, B. Xia, M. Leguia, J. C. Anderson, and D. Densmore
Eugene – a domain specific language for specifying and constraining synthetic biological parts, devices, and systems
PLoS ONE, vol. 6, iss. 4, p. e18882, 2011.
DOI:10.1371/journal.pone.0018882
L. Bilitchenko, A. Liu, and D. Densmore
The Eugene language for synthetic biology
Methods of Enzymology, vol. 498, pp. 153-172, 2011.
DOI:10.1016/B978-0-12-385120-8.00007-3
Contributors
The first version of Eugene was developed by the 2009 iGEM UC Berkeley Software Team, who also won the Best Software Tool award.
Agilent Technologies funded the development and evolution of Eugene in 2012 and 2013.
The Joint BioEnergy Institute (JBEI) integrates Eugene into the DeviceEditor CAD Tool.
DOI:10.1186/1754-1611-6-1
Life Technologies (a Thermo Fisher Scientific brand) integrates Eugene into the Vector NTI Express Designer Software.
The Boston University 2012, 2013, and 2014 iGEM Teams utilized Eugene in the design of their iGEM projects.
All members of the CIDAR Group contributed to Eugene. Specifically the following (current and affilated) supported Eugene's development:
- Evan Appleton
- Swapnil Bhatia
- Swati Carr
- Bryan Der
- Traci Haddock
- Haiyao "Cassie" Huang
- Nicholas Roehner
- Jenhan Tao
- Prashant Vaidyanathan
- Erik Lindgren
- Carter Wheatley
In Memory of

Allan Kuchinsky


